Fast DNA Replication Can Destroy the Cancer Cell

A new study led to the finding that if DNA replicates very fast it can destroy the cancer cell.
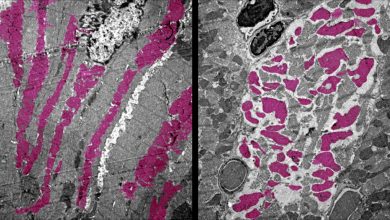
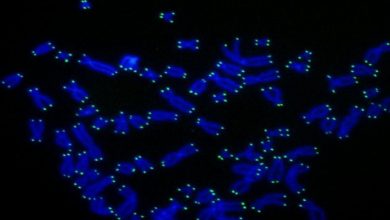
For DNA replication, the two DNA strands unwind and form a fork-like structure called replication fork. The speed of replication is determined by the molecular network involved in the regulation of replication-fork speed at which the replication fork progresses along DNA and it must be controlled for normal duplication of the genome. If it speeds up, it can cause damage to the cell.
There is a protein called PARP whose function is to repair the damaged DNA in replication fork by the process called PARylation, in which the proteins synthesize chains of ADP-ribose molecules that attract repair proteins to the damaged DNA. Through their previous studies, scientist found that PARP inhibitors drugs inhibit the PARP activity due to which replication forks collapse when encountered with DNA damage. This leads to accumulation of DNA damage owing to improper replication and death of the treated cells.
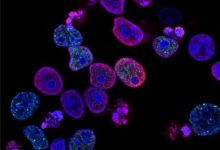
During their research, scientist treated proliferated human cells with the PARP inhibitor olaparib in vitro which caused DNA damage because the fork speed increase by 40 % due to which there was insufficient time for the forks to recognize damaged DNA which is in need of repair. Hence Damaged DNA was accumulated and the number of viable cells reduced.
The next step of this research was to find the pathway by which PARP inhibition was accelerating the replication forks. For that, Maya-Mendoza the author of the study examined protein p21, whose function is to inhibit DNA replication.
The authors found that loss of p21 led to an increase in replication-fork speed similar to the acceleration caused by PARP inhibitors. Loss of p21 in addition to PARP inhibition increased fork speed more than either manipulation in isolation.
Tumor cells having deficiencies in a pathway called homologous repair that repairs double-stranded DNA breaks are specifically susceptible to PARP inhibitors which means that when single strand with damaged DNA is replicated to ds DNA ( due to PAR inhibitors) damaged DNA is accumulated. Due to defective homologous repair pathway, the inability to repair these defects leads to cell death. Most notably, breast, ovarian and prostate cancers caused by mutations in the genes BRCA1 and BRCA2 are susceptible to PARP inhibition.
With this study in mind, researchers said that PARP inhibition accelerates fork speed above the threshold in BRCA1-deficient cells. the authors suggested that the susceptibility of tumors harboring BRCA mutations to PARP inhibitors might not be due to increased stalling and collapse of replication forks, as originally believed, but instead to the aberrant acceleration that compromises the ability of forks to detect and repair DNA damage.
The next step of the study is to the mechanism by which PARP and p21 are controlling replication fork and what others pARP are also involved.
Prominently, when PARP inhibitors were combined with chemotherapeutic agents it induces DNA damage by inhibiting the replication forks to progress. Hence, it would be amusing to determine whether the same mechanism underlies the effects of PARP inhibitors when used in combination with chemotherapy.
With this study in mind, Maya-Mendoza and colleagues’ findings will help many research groups to explore whether bettering the threshold fork speed provides a more general way to explain the molecular basis of cancer and tumor sensitivity to chemotherapy.