New Tools to Find Genes Causing Rhabdomyosarcoma-Childhood Cancer

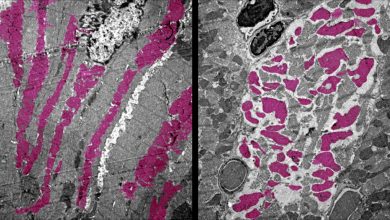
Rhabdomyosarcoma, an aggressive childhood cancer is a rare type of disorder highly malignant in nature. It is also described as small, round blue cell tumor of childhood. It occurs when sarcoma i.e. soft skeletal muscle tissue fails to differentiate completely.
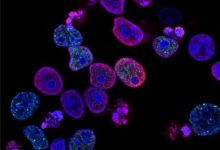
A new study by UT South Western Medical Centre led to the finding of 29 genetic changes which contributes to the Rhabdosarcoma.
The research group used Bayesian analysis which is a statistical inference and gene editing tool for CRISPR/cas9 for screening.
This research suggests that these identified gene act as an engine which leads to the formation of this Rhabdomyosarcoma and also helps to identify other cancer-causing genetic drivers.
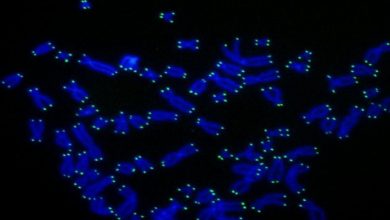
As we all know, genes always occur in pairs but when they occur as a single copy or with 2 or more copies, they lead to some disorder or disease. This research is focused on such type of genes only.
“We came up with the idea that the altered expression of key cancer genes may be driven by genomic copy-number amplifications or losses. We then developed a new computational algorithm called iExCN to predict cancer genes based on genomewide copy-number and gene expression data,” said Dr. Stephen Skapek, Chief of the Division of Pediatric Hematology-Oncology and with the Harold C. Simmons Comprehensive Cancer Center.
During their research, they used an algorithm to investigate genome data from around 209 tumors of Rhabdomyosarcoma and did the screening using CRISPR/cas9.
They identified 29 genes which were associated with Rhabdosarcoma but no link was found between them in previous studies. After that they used several new experimental tools, including CRISPR/Cas9 screening technology, to verify the function of these predicted cancer genes in rhabdomyosarcoma.
“The iExCN algorithm was developed based on Bayesian statistics, which is fundamentally different from commonly used statistics methodologies, and usually provides more accurate estimation of statistical associations, though it involves more complicated computation and longer processing time,” said Dr. Lin Xu, Instructor in the Departments of Clinical Sciences and Pediatrics and with the Quantitative Biomedical Research Center.
According to the researcher, among 29 identified genes, the genes which were needed to be further investigated are EZH2, CDK6, RIPK2 because there are already drugs that target these genes that are either FDA-approved or in clinical trials.