A new research led by Dr. Knut Woltjen reported a new gene editing technique which can transform a single DNA base in the human genome with absolute precision. The study published in nature communication describes this new method as a unique one as it guides the cell’s own repair mechanisms by design, providing pairs of genetically matched cells for studying disease-related mutations.
Single nucleotide polymorphisms, frequently called SNPs (pronounced “snips”), are the most common type of genetic variation among people. Each SNP represents a difference in a single DNA building block, called a nucleotide. More than 10 million SNPs are known, many of which are associated hereditary disease like Alzheimer’s, heart disease, and diabetes but their role in these diseases is still unknown.
In order to understand the role of SNIPs, researcher team at Kyoto University’s Center for iPS Cell Research and Application (CiRA) made induced pluripotent stem (iPS) cells from patient donors.
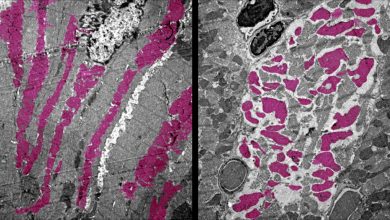
Cells from the Tissue such as the brain, heart, or pancreas can be formed from IPS cells as it retains the genetic makeup of the donor. These cells can be observed in the laboratory which can facilitate safe testing for new disease treatments before starting clinical trials. It is also difficult to verify that SNP is the source of the disease as it needs a very strict comparison to genetically matched, or isogenic, iPS cells. The ideal cells are what the researchers describe as isogenic “twins”; cells whose genomes differ only by one SNP.
However, Dr. Shin-Il Kim, a Specially-appointed Assistant Professor in the Woltjen lab and co-first author on the study, says that creating these twins is not trivial. “Usually we need to add a gene for antibiotic resistance along with the SNP to overcome low efficiency. Since that adds another change to the genome, we also need a way to remove it.”
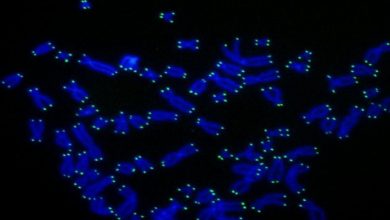
During their research, the team developed new genome editing technology for creating an isogenic twin. This technique inserts an SNP modification along with a fluorescent reporter gene which will act as a signal and can detect the transformed cells. They also engineered a short duplicated DNA sequence, known as a microhomology, on the left and right sides of the reporter gene, and unique target sites for CRISPR, an enzyme that cuts DNA. They also used microhomology-mediated end joining (MMEJ) which is a DNA repair system to remove the fluorescent reporter gene, leaving only the modified SNP behind. They generated the efficient isogenic twins by placing the mutant SNP in one microhomology and the normal SNP in the other one.
CiRA Associate Professor Dr. Knut Woltjen, called the new gene editing method MhAX, or Microhomology-Assisted eXcision. Woltjen’s inspiration came from observing naturally occurring MMEJ repair in response to DNA damage.
“To make MhAX work, we duplicate DNA sequences which are already present in the genome. We then let the cells resolve this duplication. At the same time, the cells decide which SNPs will remain after repair,” he says. “One experiment results in the full spectrum of possible SNP genotypes.”
Diabetes clinical trials using embryonic stem cells are currently underway, but chronic immune suppression is required. Gene correction of the patient’s own iPS cells could lead to a source of healthy insulin-producing pancreatic cells with a reduced chance of rejection following transplantation.
In the light of this study, the aim of the researcher is to generate gene editing technologies which can improve the understanding of the mechanism of the disease, which in turn can lead to therapies.